Clustal X
CLUSTAL X is a new windows interface for the widely-used progressive multiple sequence alignment program CLUSTAL W. The new system is easy to use, providing an integrated system for performing multiple sequence and profile alignments and analysing the results.
This application features a general purpose multiple sequence alignment program for DNA or proteins, performing comparisons and generating analysis reports

Clustal X 2.0 is the new version of the Clustal X graphical alignment tool. The original Clustal X was developed using NCBI's vibrant toolbox. The vibrant toolbox is no longer supported which led to problems compiling Clustal X on newer versions of operating systems. ClustalX is a streamlined OS X utility that provides the necessary tools to align DNA or protein sequences from within a user-friendly interface or a Terminal window. ClustalX comes with support for numerous input formats, such as GDE, FASTA, NBRF/PIR, GCG9 RSF, Clustal, GCC/MSF and EMBL/Swiss-Prot. In order to use ClustalX, you have to place. ClustalX is a streamlined OS X utility that provides the necessary tools to align DNA or protein sequences from within a user-friendly interface or a Terminal window. ClustalX comes with support for numerous input formats, such as GDE, FASTA, NBRF/PIR, GCG9 RSF, Clustal, GCC/MSF and EMBL/Swiss-Prot. In order to use ClustalX, you have to place.
Clustal X is an advanced program that deals with multiple sequence alignment for proteins and DNA. Designed as a GUI for ClustalW, the program carries out in-depth sequence analysis, while also offering many utilities to perform proper comparisons.
Load multiple specialized formats
The program is able to work with ready sequences, imported from files of the following formats: EMBL/SWISSPROT, NBRF/PIR, Pearson (Fasta), GCG/MSF (Pileup), Clustal (*.aln), GCG9/RSF and GDE.
Clustal X2

During the import process, Clustal X attempts to detect whether the sequences are nucleotides or amino acids, but there’s no actual guarantee that the choice will be correct.
Cleverly highlighting important elements
Once the source file has been loaded, the sequence alignment will be displayed in the main window of the program. The built-in coloring scheme serves to highlight conserved features, making them pop out via vivid colors.
In order to distinguish the contents of the sequences, it is recommended that you use a larger font; this will result in a zoomed sequence view, making it easier to spot its composition.
Easy alignment process
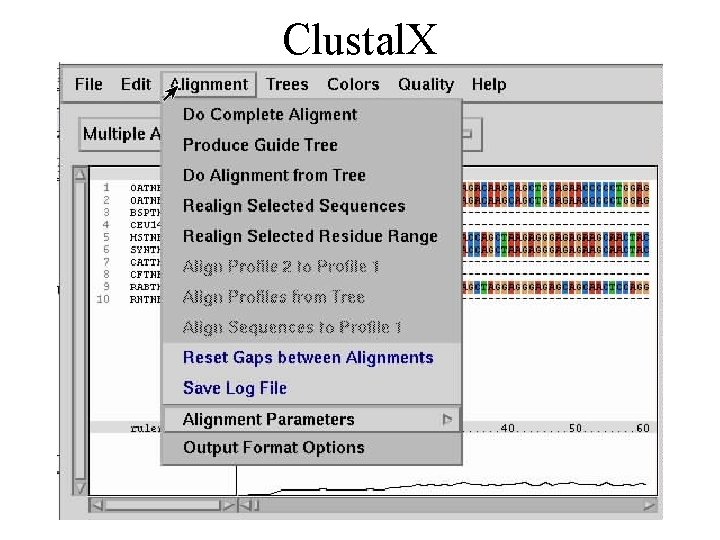
The operations allowed by the program make quite the list; you can append a sequence to an existing alignment, cut or paste a fragment, as well as to search for a string or to remove existing gaps.
Also, you can perform alignments via one of the following methods: complete (the sequence will undergo a full analysis), guide tree only or a realignment of the selected sequences and / or residue ranges.
It is recommended that before starting an alignment process, you reset all the parameters (new gaps, protein gaps) in order for the results to be conclusive.
Analyze data and get an alignment quality score
One of the most important indicative of the analysis will be the alignment quality score, which is plotted for each column individually. A high score designates a well-conserved column, while low conservation levels are indicated by a lower score.
In conclusion
However, the analysis depends on a variety of parameters, which should be set in accordance with the scientific purpose that needs to be carried out. Most definitely, the program makes a great choice for scientists, as well as students who are looking to make a carrier out of DNA sequencing.
Filed under
Clustal X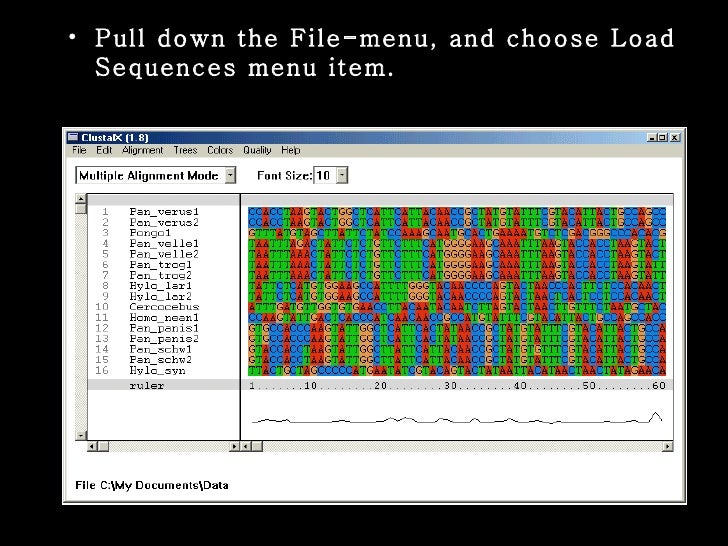 was reviewed by Andreea Matei
was reviewed by Andreea MateiClustal X 2.1
add to watchlistsend us an update- runs on:
- Windows All
- file size:
- 4.7 MB
- main category:
- Science / CAD
- developer:
- visit homepage
top alternatives FREE
top alternatives PAID

What Is Clustalx
Introduction
Clustal X is essentially the same program as Clustal W. It is a program designed to compute the alignment of multiple sequences of genetic or protein data. This program uses a GUI to work with the program as opposed to Clustal W, which just uses the command line as an interface.
Using Clustal X on RCC Resources
Clustal X, much like its sister program Clustal W, does not require a module to be loaded in order to run. Clustal X is the GUI version of Clustal W so it is primarily designed to be an interactive program. In order to start the GUI for Clustal X, simply run clustalx from the command line and the GUI will start.